Mutation Query
| | | | Allele 1: | R386C | | Allele 2: | H932Y | Allelic information known | | Refine query |
|
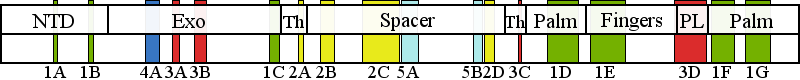
| | | Residue R386 | | Cluster assignment: | | | Cluster description: | Single Nucleotide Polymorphism | | POLG domain: | Exonuclease domain |
| Residue H932 | | Cluster assignment: | | | Cluster description: | Polymerase active site and environs | | Subcluster: | 1E (residues 914-966) | | Subcluster description: | This subcluster comprises most of the fingers subdomain of the pol domain, including the O-helix and the Pol B motif (Loh and Loeb, 2005). | | POLG domain: | Polymerase domain |
|
|
Mutation Information
|
| R386C | | | | Number of patients: (with R386C) | 1 | | Non-allelic with: | H932Y (100%) |  | Show Patient Data |
| Patient data are sorted by mutation combination frequency. | Reference: | Giordano et al, 2010; | | Description: | Isolated distal myopathy of the upper extremities, cytochrome c oxidase deficient fibers, muscle weakness. mtDNA depletion. Reduced Deep tendon reflexes in the upper extremities. | | Mutations: | H932Y | | SNPs: | R386C | | Age group: | adult | | Age of Onset: 24, Age of Patient: 27, Age of Death: n/a |
|
| H932Y | | | | Number of patients: (with H932Y) | 4 | | Found together with: | |  | Show Patient Data |
| Patient data are sorted by mutation combination frequency. | Reference: | Tang et al, 2011; | | Description: | Peripheral neuropathy, exercise intolerance, muscle cramps after exercise, easy fatigability, arrythmia, CPEO, abnormal EMG/NCV, ptosis, cataract, abnormal muscle histology, abnormal muscle ultrastructure, COX deficiency, ragged red fibers. 123% mtDNA copy number in blood. | | Mutations: | H932Y, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 31, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Peripheral neuropathy, ptosis, muscle weakness, CPK abnormalities. 77% mtDNA copy number in blood. | | Mutations: | H932Y, P587L | | SNPs: | T251I | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 41, Age of Death: n/a |
| Reference: | Mancuso et al, 2004a; | | Description: | PEO, erectile dysfunction, progressive hearing loss, dysarthria, ataxic gait, Romberg sign, severe peripheral axonal sensorimotor polyneuropathy. | | Mutations: | G1051R, H932Y | | Age group: | juvenile | | Age of Onset: 18, Age of Patient: 35, Age of Death: n/a |
| Reference: | Giordano et al, 2010; | | Description: | Isolated distal myopathy of the upper extremities, cytochrome c oxidase deficient fibers, muscle weakness. mtDNA depletion. Reduced Deep tendon reflexes in the upper extremities. | | Mutations: | H932Y | | SNPs: | R386C | | Age group: | adult | | Age of Onset: 24, Age of Patient: 27, Age of Death: n/a |
Back to top |
|
|
|
The following information is based on PON-P2 mutation pathogenicity prediction software results. Cluster 1 mutation with a non-cluster-mapping mutation (SNP) R386C | PON-P2 predictions results for R386C | | Pathogenicity: | Pathogenic | | Probability: | 0.799 | | Standard Error: | 0.066 | | Prediction result is based on sequence analysis only and may not be accurate. |
Risk of POLG-related syndromes exists.
Mutation pathogenicity prediction suggests that this mutation could be pathogenic. Predicted chance of pathogenicity is 79.9%. See further details for residue 386. All mutations mapping into the pathogenic clusters are in high risk of being pathogenic. As a rule, a patient must have a pathogenic mutation in both of his/ her POLG genes to develop a POLG-related syndrome. |
|