Mutation Query
| | | | Allele 1: | G517V | | Allele 2: | R1128H | Allelic information known | | Refine query |
|
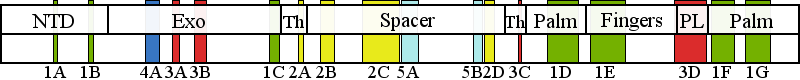
| | | Residue G517 | | Cluster assignment: | | | Cluster description: | Upstream DNA binding channel | | Subcluster: | 2B (residues 496-517) | | Subcluster description: | Subcluster 2B maps to the region of the AID that is predicted to contact the upstream DNA duplex. | | POLG domain: | Spacer domain |
| Residue R1128 | | Cluster assignment: | | | Cluster description: | Polymerase active site and environs | | Subcluster: | 1F (residues 1098-1138) | | Subcluster description: | This subcluster forms a segment of the pol active site and contains two highly conserved motifs that are found in all family A polymerases, the Pol C motif and motif 6 (Loh and Loeb, 2005). | | POLG domain: | Polymerase domain |
|
|
Mutation Information
|
| G517V | | | | Number of patients: (with G517V) | 12 | | Found as the only mutation: | 67% of entries (8 patients) | | Found together with: | |  | Show Patient Data |
| Patient data are sorted by mutation combination frequency. | Reference: | Burusnukul and de los Reyes, 2009; | | Description: | Global developmental delay, Central hypotonia, Nystagmus and eye surgery, Autistic features, Generalized seizure, Hypnic myoclonia, Intermittent hypoglycemia, Abnormal MRI | | Mutations: | G517V | | Age group: | infantile | | Age of Onset: 0.7, Age of Patient: 4.5, Age of Death: n/a |
| Reference: | Burusnukul and de los Reyes, 2009; | | Description: | Migraines, Complex partial seizure with secondarily generalization, mild developmental delay, muscle cramps and experienced easy fatigability, | | Mutations: | G517V | | Age group: | childhood | | Age of Onset: 7, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Hopkins et al, 2010; | | Description: | diagnosed with type I DM at age 2, diagnosed with adrenal insufficiency and hypothyroidism at age 10, first seizure or dystonic crisis at age 16 years, then neurologic, intensive care unit, and psychiatric admissions for dystonic crises, status epilepticus, nonconvulsive status epilepticus, encephalopathy, depression, suicidal ideation, and psychosis. Elevated lactate, chronic myopathic changes, developmental delay | | Mutations: | G517V | | Age group: | infantile | | Age of Onset: 2, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Hopkins et al, 2010; | | Description: | diagnosed with type I DM at age 5, diagnosed with adrenal insufficiency and hypothyroidism at age 11, presented to neurology at age 17 years, for 2 weeks of headache and recent onset of left facial twitching. neurologic, intensive care unit, and psychiatric admissions for dystonic crises, status epilepticus, nonconvulsive status epilepticus, encephalopathy, depression, suicidal ideation, and psychosis. Elevated lactate, developmental delay | | Mutations: | G517V | | Age group: | childhood | | Age of Onset: 5, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Woodbridge et al, 2012; | | Description: | neuropathy, dysphagia/ dysarthria, slowly progressive ataxia, hearing loss, slow gastrointestinal transit times, proximal myopathy. Patient #5. | | Mutations: | G517V | | Age group: | juvenile | | Age of Onset: n/a, Age of Patient: 56, Age of Death: n/a |
| Reference: | Staropoli et al, 2012; | | Description: | Diffuse hypotonia, hypoactive reflexes, and roving eye movements. Decreased vis- ual acuity, mild bilateral macular pigmentary changes, normal refractive indices, bilateral ptosis, diffuse cerebral atrophy, and disconjugate, nystagmoid eye movements. In addition to G517V, affected by gene CLN5 c.61C>T, p.Pro21Ser and de novo exon 3 deletion. | | Mutations: | G517V | | Age group: | infantile | | Age of Onset: 0.1, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Schicks et al, 2010; | | Description: | PEO, late-onset ataxia, dysarthria, ptosis, pyramidal tract signs, psychiatric disorders | | Mutations: | G517V | | Age group: | adult | | Age of Onset: 44, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Schicks et al, 2010; | | Description: | early-onset ataxia, dysarthria, pyramidal tract signs, psychiatric disorders, Cerebellar atrophy, Extrapyramidal signs. | | Mutations: | G517V | | Age group: | adult | | Age of Onset: 22, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Wong et al, 2008; | | Description: | Onset at 1 years with microcephaly, developmental delay/ dementia, and liver dysfunction. Authors reported as undiagnosed. | | Mutations: | G517V, R1128H | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: n/a |
Back to top | Reference: | Wong et al, 2008; | | Description: | Onset at 1 years with myopathy, developmental delay/ dementia, RRF, and elevated CK. Authors reported as undiagnosed. | | Mutations: | D1196N, G517V | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: n/a |
| Reference: | Bolszak et al, 2009; | | Description: | Onset at 4 months with generalized tonic-clonic seizure, evolved to status epilepticus, treated with valproate, Progressive encephalopathy, and psychomotor development deficient by age 2, mentally retarded, ataxic and hyperkinetic. At the last follow-up visit at age 17 years, he was severely retarded, autistic, and ataxic. During 12 months of valproate treatment serum alanine aminotransferase (ALAT) increased from 29 to 71 U/L (normal < 40 U/L), and after discontinuation of the medication, ALAT values varied between 5 and 12 U/L. | | Mutations: | G517V | | SNPs: | R722H | | Age group: | infantile | | Age of Onset: 0.33, Age of Patient: 17, Age of Death: n/a |
| Reference: | Tang et al, 2011; | | Description: | Peripheral neuropathy, myopathy, ptosis, CPEO, COX deficiency, ragged red fibres | | Mutations: | G517V, Y955C | | Age group: | adult | | Age of Onset: n/a, Age of Patient: 47, Age of Death: n/a |
|
| R1128H | | | | Number of patients: (with R1128H) | 1 | | Non-allelic with: | G517V (100%) |  | Show Patient Data |
| Patient data are sorted by mutation combination frequency. | Reference: | Wong et al, 2008; | | Description: | Onset at 1 years with microcephaly, developmental delay/ dementia, and liver dysfunction. Authors reported as undiagnosed. | | Mutations: | G517V, R1128H | | Age group: | infantile | | Age of Onset: 1, Age of Patient: n/a, Age of Death: n/a |
|
|
|
|
|
The following information is based on existing patient data and pathogenic cluster assignment.
Pathogenicity information for a patient with mutations in Clusters 1 and 2: Age of onset information is extracted from a total of 93 patients and/ or patient families. | Age of onset | | |
93- 47- | 57
| 13
| 2
| 21
| | | infantile | childhd | juvenile | adult | | | 61% | 14% | 2% | 23% | |
All mutations mapping within the pathogenic clusters are at high risk for pathogenicity. In general, a patient must have a pathogenic mutation in both of his/ her POLG genes to develop a POLG-related syndrome.  | Symptoms described in patients with cluster2-cluster1 mutations | |
| Symptoms in patients with combination
cluster1:cluster2 | | Encephalopathy | 43.8% | | Developmental delay | 43.8% | | Epilepsy | 35.4% | | Movement disorder (ataxia) | 19.8% | | PEO | 19.8% | | Dementia | 18.8% | | Ptosis | 17.7% | | Hypotonic | 16.7% | | Lactic acidosis | 14.6% | | Liver failure | 12.5% | | Myoclonic seizures | 10.4% | | Epilepsia partialis | 10.4% | | Peripheral neuropathy | 9.4% | | Failure to thrive | 9.4% | | Ragged red fibers | 8.3% | | Muscle weakness | 8.3% | | Liver dysfunction | 8.3% | | Status epilepticus | 7.3% | | Intractable seizure | 7.3% | | Myopathy | 7.3% | | Hypoglycemia | 7.3% | | Tremor | 6.3% | | Stroke | 5.2% | | +44 other symptoms in under 5.0% of the patients |
| | Data gathered from clinical descriptions for 96 patients |
| Symptoms by group | | Developmental Delay | 58.3% | | Seizures | 58.3% | | Alpers syndrome | 33.3% | | Hepatopathy | 30.2% | | CPEO | 27.1% | | Other | 22.9% | | Myopathy | 20.8% | | Ataxia | 19.8% | | Hypotonia | 16.7% | | Neuropathy | 15.6% | | CNS symptoms | 14.6% | | GI symptoms | 8.3% | | Migraines | 3.1% | | Unknown | 3.1% |
| | [Show grouping information] |
|
|
|