| Home | Query | References | Browse | Contact |
Pathogenic clusters on DNA Polymerase gamma primary amino acid sequence.
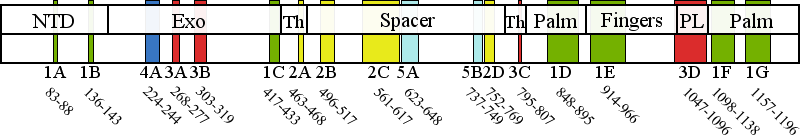
Pathogenic cluster definitions in PolG primary amino acid sequence.
| Cluster | Sub- cluster | Residue range | Description |
Cluster 1 Polymerase active site and environs | 1A | 83-88 | No biochemical data are available for this region. We predict an architectural role via stabilization of subcluster 1G. |
| 1B | 136-143 | No biochemical data are available for this region. We predict an architectural role via stabilization of subclusters 1C, 1G, and 1F. | |
| 1C | 417-433 | No biochemical data are available for this region. This subcluster is adjacent to motif 2 and motif 6 in the pol domain, and we propose it serves an architectural role that contributes to pol activity indirectly, by stabilizing subclusters 1B, 1D, 1F, and 1G. Patient data shows these mutations to be recessive. | |
| 1D | 848-895 | Motif 2 and the Pol A motif are both critical for catalysis but contribute to catalysis by different mechanisms. Therefore, a mutation in subcluster 1D should be evaluated by the motif to which it is closest in primary sequence. Motif 2 (residues 845-863) In PolG, motif 2 in the pol domain has been termed the "RR loop" because of two sequential arginines (R852 and R853) located on the tip of the loop, whereas family A polymerases have only a single arginine (equivalent to R853) (Euro et al, 2011). Overall, motif 2 is critical for binding correctly base-paired primer-template DNA in the minor groove, and to stabilize the template DNA backbone in the active site (Loh and Loeb, 2005). These residues form specific contacts with DNA in the pol active site and are major determinants for the catalytic efficiency and fidelity of DNA synthesis by Pol ?. Motif 2 mutations should always cause reduced pol activity but may also cause a DNA binding defect depending on the residue, such as the 5-fold defects reported for G848S and R852C (Kasiviswanathan et al, 2009). The DNA binding affinity of T851A, R853Q, Q879H, T885S may be decreased slightly, but the reduction in pol activity is much greater (Kasiviswanathan et al, 2009). Overall, it is unlikely for motif 2 mutations to present as dominant mutations because these mutant forms of Pol ? would not compete effectively with wild type Pol ? for DNA binding. Pol A motif (residues 887-896) The Pol A motif of family A DNA polymerases forms the site of catalysis where it contacts closely the primer strand and binds the Mg2+ ion required for the chemical steps of nucleotide polymerization (D890 in PolG). Additionally, a residue of the Pol A motif discriminates against ribonucleotide incorporation (E895 in Pol ?), and mutation causes severe pol defects in addition to increased ribonucleotide incorporation (Astatke et al, 1998). Mutations in the Pol A motif at either D890 or E895 in Pol ? would likely manifest a dominant lethal phenotype, and the only patient carrying the E895G mutation to date died immediately after birth (Spinazzola et al, 2009). In contrast to these two critical residues, adjacent residues within the Pol A motif play lesser roles in pol activity and mutations in them are observed more frequently, resulting in reduced pol activity without a significant effect on DNA binding affinity, and thus they can produce a dominant phenotype. Overall, though dominant mutations are fairly rare for subcluster 1D, novel mutations affecting residues of the Pol A motif would present a moderate risk for dominant inheritance. | |
| 1E | 914-966 | Pol B motif (residues 943-958) The conserved amino acid residues on the O-helix are known as the Pol B motif, and function to establish specific contacts with correctly base paired dNTP in the closed conformation. POLG mutations that disrupt the specific contacts with the incoming dNTP (H932Y, R943H, K947R, Y951N and Y955C) will reduce fidelity and increase Km (dNTP), without affecting DNA binding affinity (Graziewicz et al, 2004; Batabyal et al, 2010). Mutations affecting this site will most likely be dominant because they are capable of competing for dNTP binding with wild type Pol ?, but are unable to polymerize nucleotides effectively, and are predicted to cause mtDNA damage that is associated with enzyme stalling (Atanassova et al, 2011). | |
| 1F | 1098-1138 | Motif 6 (residues 1097-1110) is located on the Q-helix and binds correctly base paired template strand and the minor groove of the primer-template DNA. The Pol C motif (residues 1134-1141) binds Mg2+ via residue D1135, whereas H1134 and E1136 contact the primer strand. Mutations in the Pol C motif will affect the rate of the chemical step of dNTP addition to the primer terminus. | |
| 1G | 1157-1196 | This subcluster is adjacent to motifs A and C, and we propose it serves an architectural role that contributes to pol activity indirectly, by stabilizing subclusters 1D and 1F. Only K1191 is predicted to contact the primer terminus, whereas the remainder of the subcluster 1G residues serve an architectural role. Patient data shows these mutations to be recessive. | |
Cluster 2 Upstream DNA binding channel | 2A | 463-468 | Extensive biochemical analysis of the spacer domain in general has demonstrated that the hydrophobic core of the IP subdomain is critical for shaping the DNA binding channel wall, and mutations are predicted to perturb the channel to block DNA from entering, which results in reduced DNA binding affinity. |
| 2B | 496-517 | The AID is stabilized in the presence of the accessory subunit and enhances DNA binding affinity through many positively charged residues. Subcluster 2B maps to the region of the AID containing the positively charged residues, and mutations may alter their interaction with bound DNA. | |
| 2C | 561-617 | Extensive biochemical analysis of the spacer domain in general has demonstrated that the hydrophobic core of the IP subdomain is critical for shaping the DNA binding channel wall, and mutations are predicted to perturb the channel to block DNA from entering, which is then observed as reduced DNA binding affinity. | |
| 2D | 752-769 | Extensive biochemical analysis of the spacer domain in general has demonstrated that the hydrophobic core of the IP subdomain is critical for shaping the DNA binding channel wall, and mutations are predicted to perturb the channel to block DNA from entering, which is then observed as reduced DNA binding affinity. | |
Cluster 3 Partitioning loop | 3A | 268-277 | The Exo II motif has been demonstrated to alter the pol: exo activity ratio in biochemical variants of Pol I by decreasing the affinity of the primer strand for the exo active site, consistent with the decrease in exo activity and wild type pol activity observed for the R233W variant in yeast Pol ? (H277 in human PolG) (Foury and Szczepanowska, 2011). |
| 3B | 303-319 | All biochemical variants of the orienter module have shown reduced DNA-binding affinity and reduced pol activity and in addition, the L304R variant showed a significant increase in exo activity (Foury and Szczepanowska, 2010). | |
| 3C | 795-807 | Mutations in subcluster 3C have been reported to cause DNA binding defects in most biochemical variants studied. Notably, an SYW triple alanine substitution in recombinant fly PolG, which was found to exhibit increased exo activity with both decreased pol activity and DNA binding affinity (Luo and Kaguni, 2005), maps adjacent to subcluster 3C in human PolG (residues 799SFW801). Biochemical study of a variant of yeast PolG M602I (R802 in Hs PolG) showed wild type DNA binding affinity, decreased pol activity, and increased exo activity (Foury and Szczepanowska, 2011). | |
| 3D | 1047-1096 | To date, no biochemical data are available for residues of the partitioning loop, although it has been shown that yeast strains homozygous for a mutation equivalent to G1051R in human PolG cause a 10-fold increase in point mutational frequency in vivo (Baruffini et al, 2007). | |
Cluster 4 Distal accessory subunit interface | 4A | 224-244 | Biochemical studies of a human R232G variant in reconstituted holoenzyme form showed a decrease in pol rate and an increase in exo activity, with unchanged DNA binding affinity that derives from loss of a direct stimulation of pol activity by the distal accessory subunit (Lee et al, 2010). We propose that the other residues of cluster 4 will have similar biochemical characteristics. |
Cluster 5 Putative protein-protein interactions | 5A | 623-648 | Cluster 5 mutations are located in the periphery of the IP subdomain and have not been shown to cause a biochemical defect in PolG per se. We predict that these mutations may affect as yet undetermined protein-protein interactions in vivo (Euro et al, 2011). |
| 5B | 737-749 | Cluster 5 mutations are located in the periphery of the IP subdomain and have not been shown to cause a biochemical defect in PolG per se. We predict that these mutations may affect as yet undetermined protein-protein interactions in vivo (Euro et al, 2011). | |
Patient data references for cluster combinations
Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 | Cluster 5 | None | |
Cluster 1 | 20 | 96 | 18 | 5 | 51 | 68 |
Cluster 2 | 69 | 33 | 8 | 60 | 34 | |
Cluster 3 | 21 | 0 | 15 | 18 | ||
Cluster 4 | 0 | 4 | 3 | |||
Cluster 5 | 70 | 7 |